FAQ¶
Q: How can I use the Dask diagnostics (bokeh) dashboard?¶
Dask distributed features a nice web interface for monitoring the execution of a Dask computation graph.
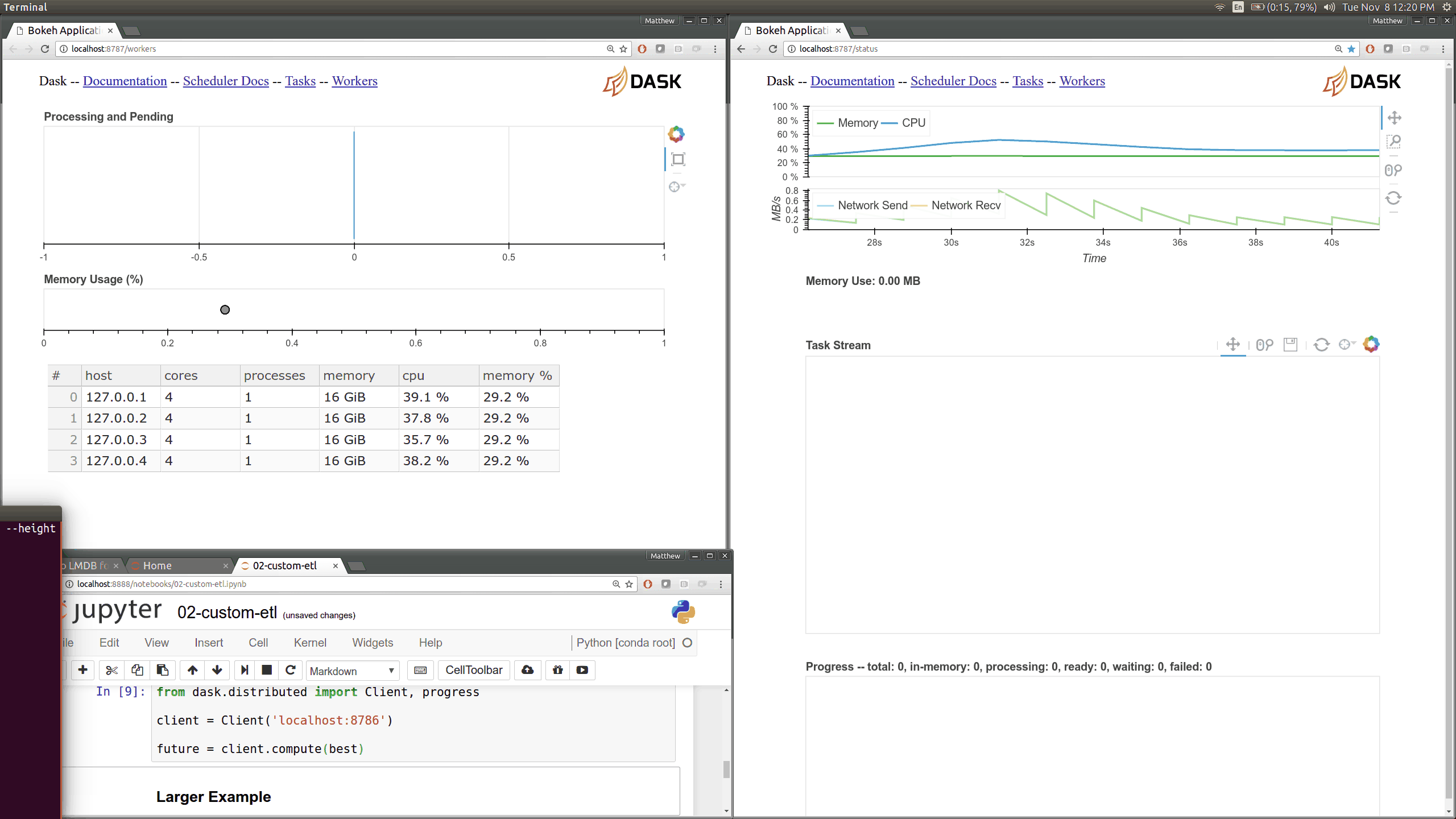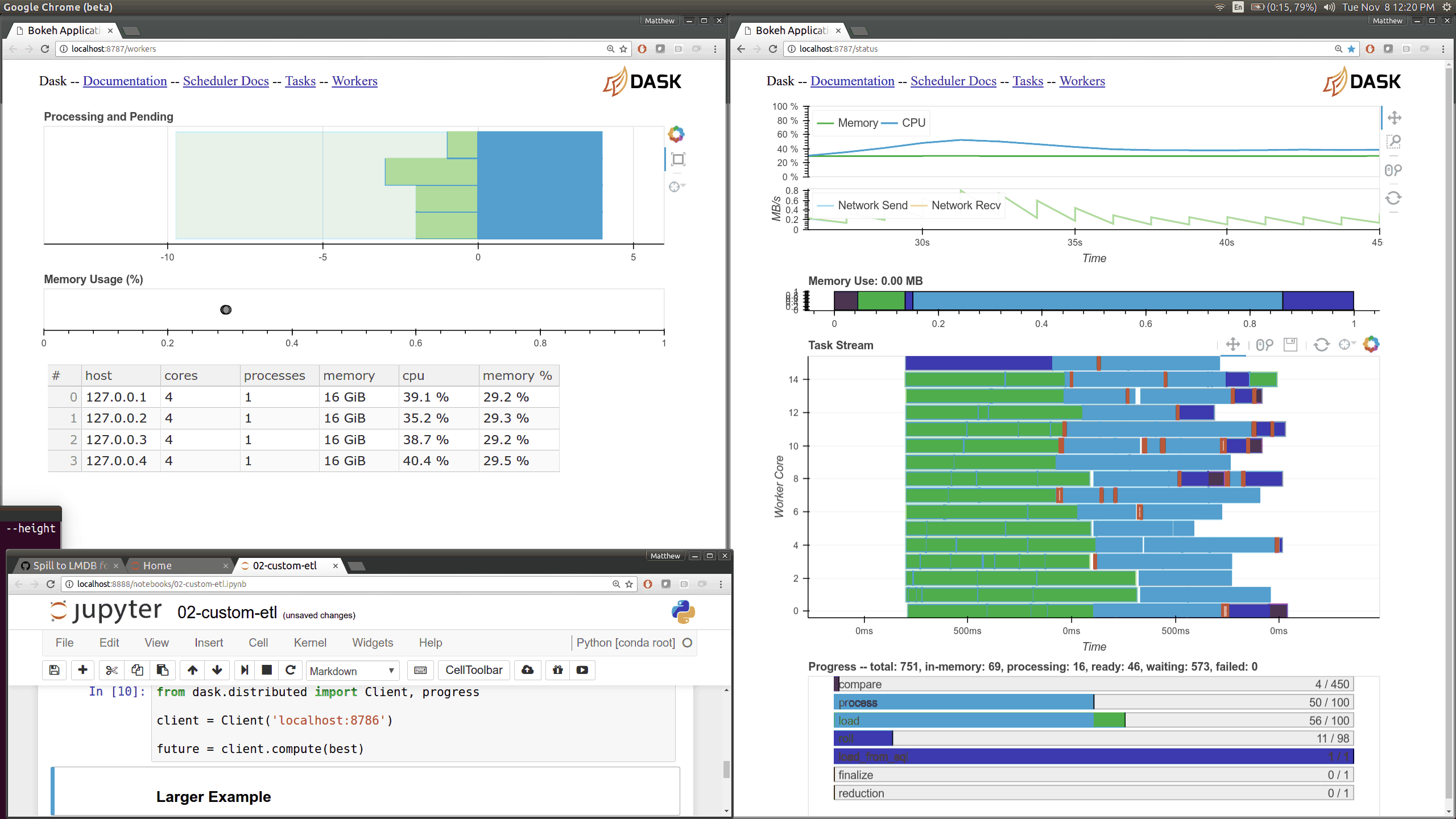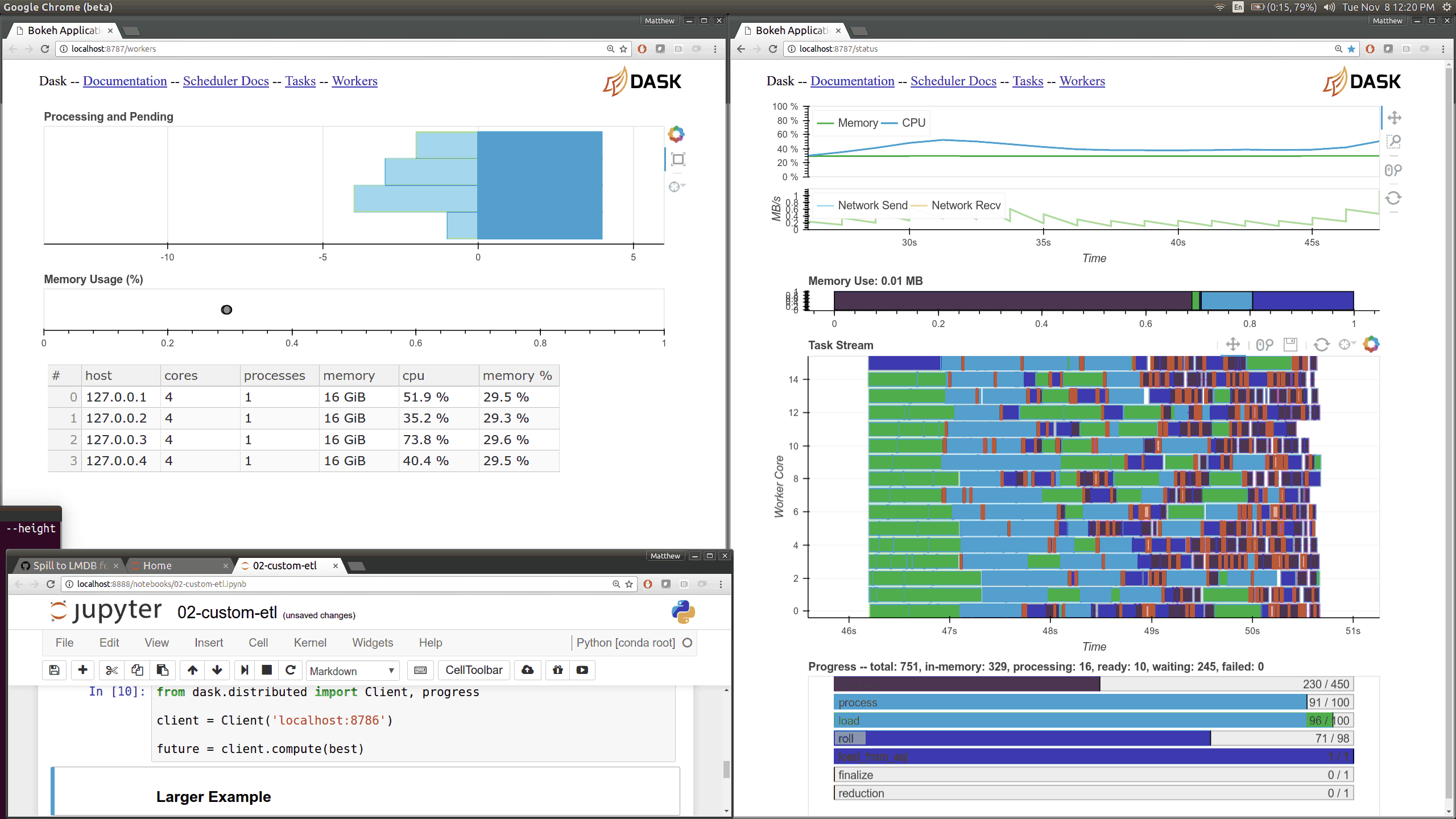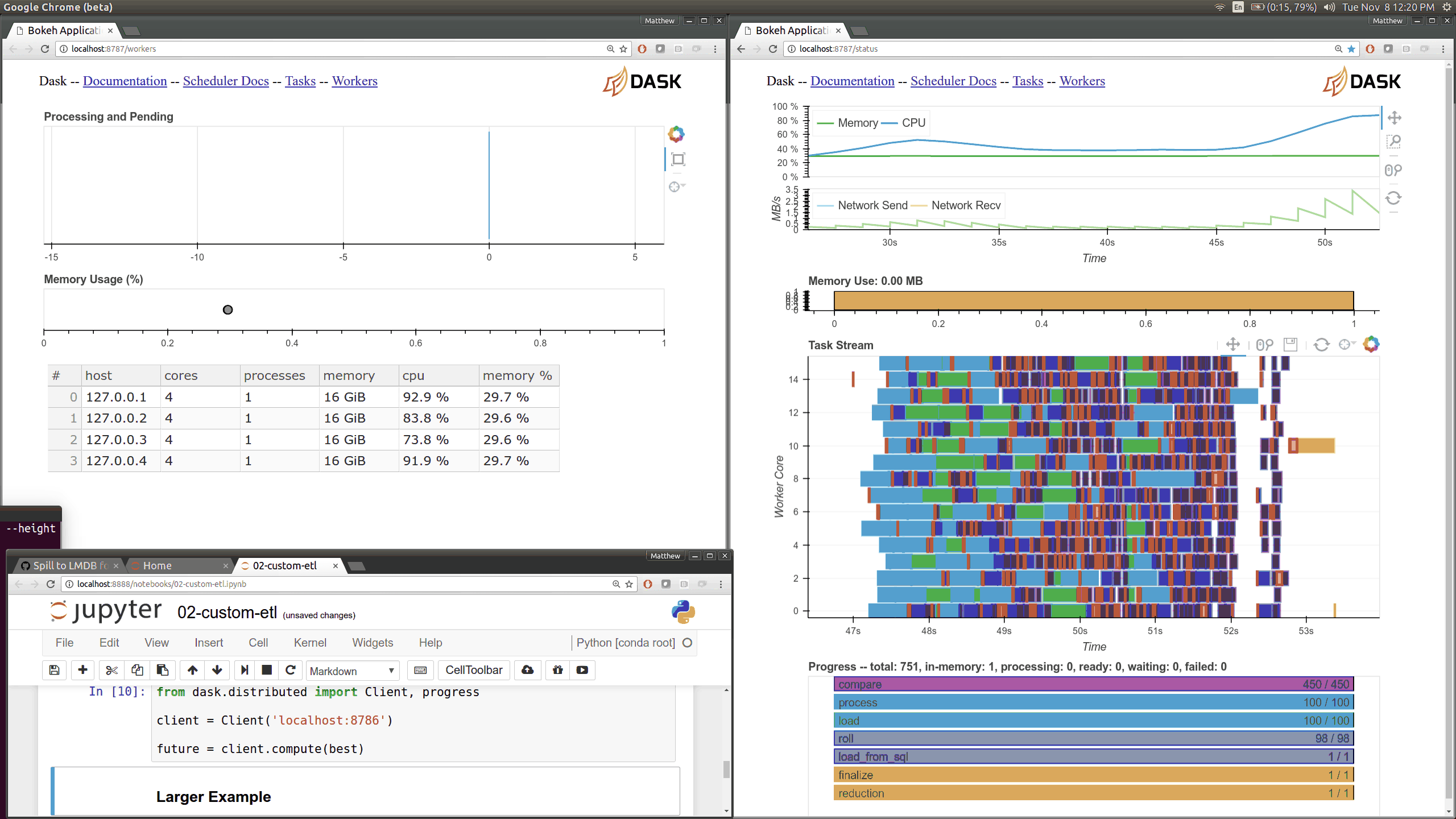
By default, when no custom Client is specified, Arboreto creates a LocalCluster instance with the diagnostics dashboard disabled:
...
local_cluster = LocalCluster(diagnostics_port=None)
client = Client(local_cluster)
...
You can easily create a custom LocalCluster, with the dashboard enabled, and pass a custom Client connected to that cluster to the GRN inference algorithm:
local_cluster = LocalCluster() # diagnostics dashboard is enabled
custom_client = Client(local_cluster)
...
network = grnboost2(expression_data=ex_matrix,
tf_names=tf_names,
client=custom_client) # specify the custom client
By default, the dashboard is available on port 8787.
For more information, consult:
- Dask web interface documentation
- Running with a custom Dask Client
Q: My gene expression matrix is transposed, what now?¶
The Python scikit-learn library expects data in a format where rows represent observations and columns represent features (in our case: genes), for example, see the GradientBoostingRegressor API.
However, in some fields (like single-cell genomics), the default is inversed: the rows represent genes and the columns represent the observations.
In order to maintain an API that is as lean is possible, Arboreto adopts the scikit-learn convention (rows=observations, columns=features). This means that the user is responsible for providing the data in the right shape.
Fortunately, the Pandas and Numpy libraries feature all the necessary functions to preprocess your data.
Example: reading a transposed text file with Pandas¶
df = pd.read_csv(<ex_path>, index_col=0, sep='\t').T
Caution
Don’t carelessly copy/paste above snippet. Take into account absence or presence of 1 or multiple header lines in the file.
Always check whether the your DataFrame has the expected dimensions!
In[10]: df.shape
Out[10]: (17650, 14086) # example
Q: Different runs produce different network outputs, why?¶
Both GENIE3 and GRNBoost2 are based on stochastic machine learning techniques, which use a random number generator internally to perform random sub-sampling of observations and features when building decision trees.
To stabilize the output, Arboreto accepts a seed value that is used to initialize the random number generator used by the machine learning algorithms.
network_df = grnboost2(expression_data=ex_matrix,
tf_names=tf_names,
seed=777)
Troubleshooting¶
Bokeh error when launching Dask scheduler¶
vsc12345@r6i0n5 ~ 12:00 $ dask-scheduler
distributed.scheduler - INFO - -----------------------------------------------
distributed.scheduler - INFO - Could not launch service: ('bokeh', 8787)
Traceback (most recent call last):
File "/data/leuven/software/biomed/Anaconda/5-Python-3.6/lib/python3.6/site-packages/distributed/scheduler.py", line 430, in start_services
service.listen((listen_ip, port))
File "/data/leuven/software/biomed/Anaconda/5-Python-3.6/lib/python3.6/site-packages/distributed/bokeh/core.py", line 31, in listen
**kwargs)
File "/data/leuven/software/biomed/Anaconda/5-Python-3.6/lib/python3.6/site-packages/bokeh/server/server.py", line 371, in __init__
tornado_app = BokehTornado(applications, extra_websocket_origins=extra_websocket_origins, prefix=self.prefix, **kwargs)
TypeError: __init__() got an unexpected keyword argument 'host'
distributed.scheduler - INFO - Scheduler at: tcp://10.118.224.134:8786
distributed.scheduler - INFO - http at: :9786
distributed.scheduler - INFO - Local Directory: /tmp/scheduler-y6b8mnih
distributed.scheduler - INFO - -----------------------------------------------
distributed.scheduler - INFO - Receive client connection: Client-7b476bf6-c6d8-11e7-b839-a0040220fe80
distributed.scheduler - INFO - End scheduler at 'tcp://:8786'
- known error: see Github issue (closed), fixed in Dask.distributed version
0.20.0 - workaround: launch with bokeh disabled:
dask-scheduler --no-bokeh - solution: upgrade to Dask distributed
0.20.0or higher
Workers do not connect with Dask scheduler¶
We have observed that sometimes when running the dask-worker command, the
workers start but no connections are made to the scheduler.
Solutions:
- delete the
dask-worker-spacedirectory before starting the workers. - specifying the
local_dir(with enough space) when instantiating a Dask
distributed Client:
>>> from dask.distributed import Client, LocalCluster
>>> worker_kwargs = {'local_dir': '/tmp'}
>>> cluster = LocalCluster(**worker_kwargs)
>>> client = Client(cluster)
>>> client
<Client: scheduler='tcp://127.0.0.1:41803' processes=28 cores=28>
- Github issue: https://github.com/dask/distributed/issues/1707