User Guide¶
Modules overview¶
Arboreto consists of multiple python modules:
arboreto.algo¶
arboreto.core¶
- Intended for advanced users.
- Contains the low-level building blocks of the Arboreto framework.
arboreto.utils¶
- Contains small utility functions.
Dependencies Overview¶
Arboreto uses well-established libraries from the Python ecosystem. Arboreto avoids being a proverbial “batteries-included” library, as such an approach often entails unnecessary complexity and maintenance. Arboreto aims at doing only one thing, and doing it well.
Concretely, the user will be exposed to one or more of following dependencies:
- Pandas or NumPy: the user is expected to provide the input data in an expected format. Pandas and NumPy are well equipped with functions for data preprocessing.
- Dask.distributed: to run Arboreto on a cluster, the user is responsible for setting up a network of a scheduler and workers.
- scikit-learn: relevant for advanced users only. Arboreto can run “DIY” inference where the user provides their own parameters for the Random Forest or Gradient Boosting regressors.
Input / Output¶
INPUT
- a list of gene names corresponding to the columns of the expression matrix
- optional
- a list of transcription factors (a.k.a. TFs)
- optional
OUTPUT
Tip
As data for following code snippets, you can use the data for network 1 from the DREAM5 challenge (included in the resources folder of the Github repository):
<ex_path>= net1_expression_data.tsv<tf_path>= net1_transcription_factors.tsv
Expression matrix as a Pandas DataFrame¶
The input can be specified in a number of ways. Arguably the most straightforward way is to specify the expression matrix as a Pandas DataFrame, which also contains the gene names as the column header.
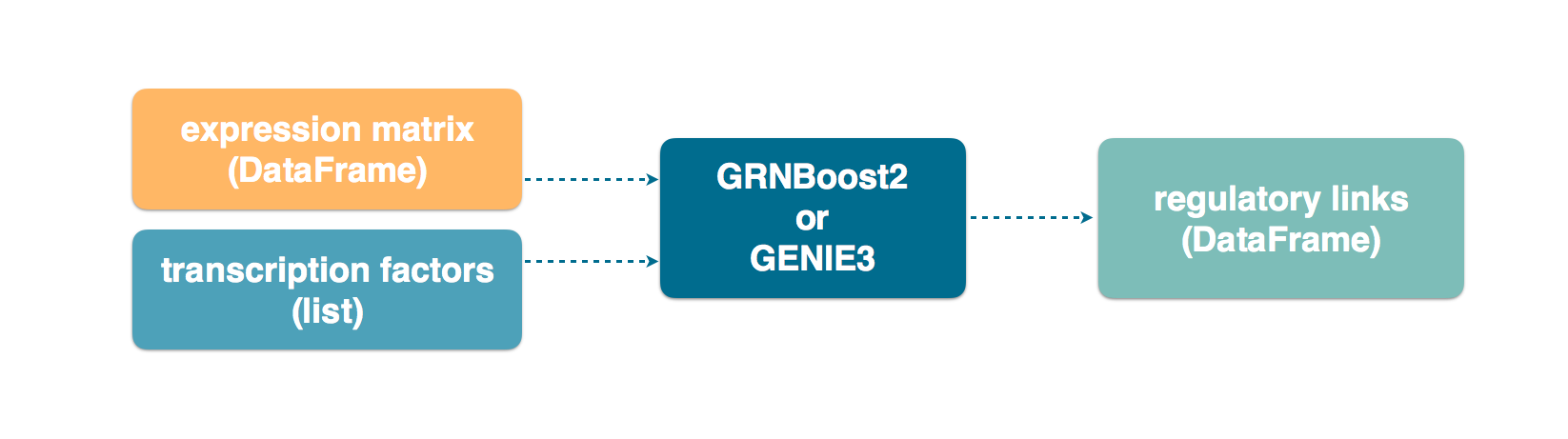
In the following code snippet, we launch network inference with grnboost2 by
specifying the expression_data as a DataFrame.
import pandas as pd
from arboreto.utils import load_tf_names
from arboreto.algo import grnboost2
if __name__ == '__main__':
# ex_matrix is a DataFrame with gene names as column names
ex_matrix = pd.read_csv(<ex_path>, sep='\t')
# tf_names is read using a utility function included in Arboreto
tf_names = load_tf_names(<tf_path>)
network = grnboost2(expression_data=ex_matrix,
tf_names=tf_names)
network.to_csv('output.tsv', sep='\t', index=False, header=False)
Note
Notice the emphasized line:
if __name__ == '__main__':
# ... code ...
This is a Python idiom necessary in situations where the code spawns new
Python processes, which Dask does under the hood of the grnboost2 and
genie3 functions to parallelize the workload.
Expression matrix as a NumPy ndarray¶
Arboreto also supports specifying the expression matrix as a Numpy ndarray (in our case: a 2-dimensional matrix). In this case, the gene names must be specified explicitly.
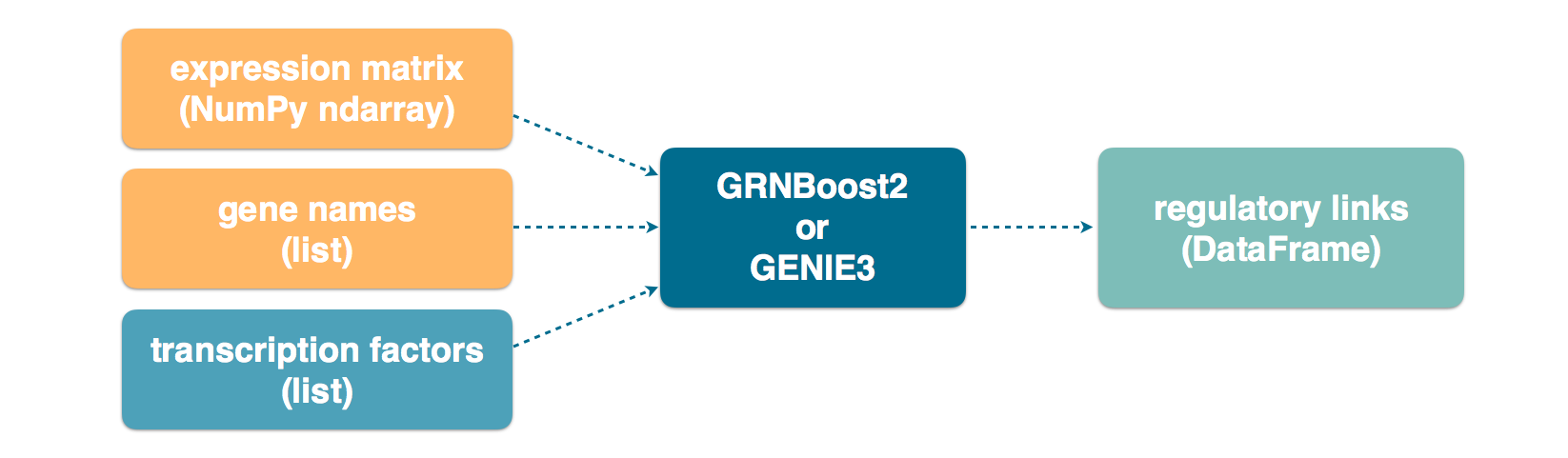
Caution
You must specify the gene names in the same order as their corresponding columns of the NumPy matrix. Getting this right is the user’s responsibility.
import numpy as np
from arboreto.utils import load_tf_names
from arboreto.algo import grnboost2
if __name__ == '__main__':
# ex_matrix is a numpy ndarray, which has no notion of column names
ex_matrix = np.genfromtxt(<ex_path>, delimiter='\t', skip_header=1)
# we read the gene names from the first line of the file
with open(<ex_path>) as file:
gene_names = [gene.strip() for gene in file.readline().split('\t')]
# sanity check to verify the ndarray's nr of columns equals the length of the gene_names list
assert ex_matrix.shape[1] == len(gene_names)
# tf_names is read using a utility function included in Arboreto
tf_names = load_tf_names(<tf_path>)
network = grnboost2(expression_data=ex_matrix,
gene_names=gene_names, # specify the gene_names
tf_names=tf_names)
network.to_csv('output.tsv', sep='\t', index=False, header=False)
Running with a custom Dask Client¶
Arboreto uses Dask.distributed to parallelize its workloads. When the user doesn’t specify a dask distributed Client explicitly, Arboreto will create a LocalCluster and a Client pointing to it.
Alternatively, you can create and configure your own Client instance and pass it on to Arboreto. Situations where this is useful include:
- inferring multiple networks from different datasets
- inferring multiple networks using different parameters from the same dataset
- the user requires custom configuration for the LocalCluster (memory limit, nr of processes, etc.)
Following snippet illustrates running the gene regulatory network inference multiple times, with different initialization seed values. We create one Client and pass it to the different inference steps.
import pandas as pd
from arboreto.utils import load_tf_names
from arboreto.algo import grnboost2
from distributed import LocalCluster, Client
if __name__ == '__main__':
# create custom LocalCluster and Client instances
local_cluster = LocalCluster(n_workers=10,
threads_per_worker=1,
memory_limit=8e9)
custom_client = Client(local_cluster)
# load the data
ex_matrix = pd.read_csv(<ex_path>, sep='\t')
tf_names = load_tf_names(<tf_path>)
# run GRN inference multiple times
network_666 = grnboost2(expression_data=ex_matrix,
tf_names=tf_names,
client_or_address=custom_client, # specify the custom client
seed=666)
network_777 = grnboost2(expression_data=ex_matrix,
tf_names=tf_names,
client_or_address=custom_client, # specify the custom client
seed=777)
# close the Client and LocalCluster after use
client.close()
local_cluster.close()
network_666.to_csv('output_666.tsv', sep='\t', index=False, header=False)
network_777.to_csv('output_777.tsv', sep='\t', index=False, header=False)
Running with a Dask distributed scheduler¶
Arboreto was designed to run gene regulatory network inference in a distributed
setting. In distributed mode, some effort by the user or a systems administrator
is required to set up a dask.distributed scheduler and some workers.
Tip
Please refer to the Dask distributed network setup documentation for instructions on how to set up a Dask distributed cluster.
Following diagram illustrates a possible topology of a Dask distributed cluster.
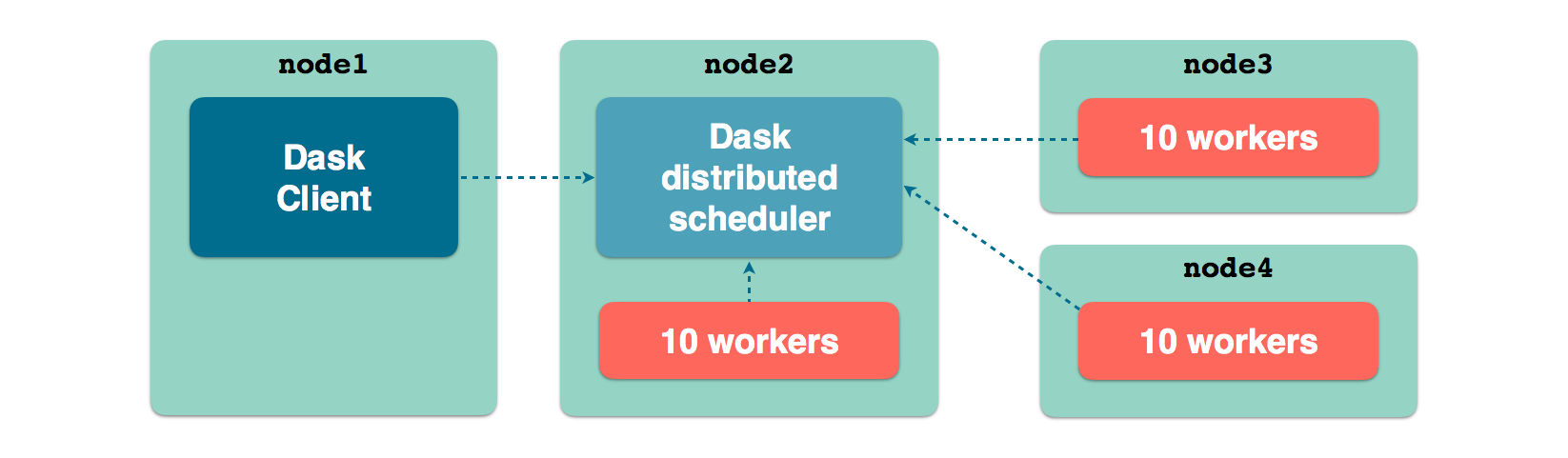
node_1runs a Python script, console or a Jupyter notebook server, a Client instance is configured with the TCP address of the distributed scheduler, running onnode_2node_2runs a distributed scheduler and 10 workers pointing to the schedulernode_3runs 10 distributed workers pointing to the schedulernode_4runs 10 distributed workers pointing to the scheduler
With a small modification to the code, we can infer a regulatory network using all workers connected to the distributed scheduler. We specify a Client that is connected to the Dask distributed scheduler and pass it as an argument to the inference function.
import pandas as pd
from arboreto.utils import load_tf_names
from arboreto.algo import grnboost2
from distributed import Client
if __name__ == '__main__':
ex_matrix = pd.read_csv(<ex_path>, sep='\t')
tf_names = load_tf_names(<tf_path>)
scheduler_address = 'tcp://10.118.224.134:8786' # example address of the remote scheduler
cluster_client = Client(scheduler_address) # create a custom Client
network = grnboost2(expression_data=ex_matrix,
tf_names=tf_names,
client_or_address=cluster_client) # specify Client connected to the remote scheduler
network.to_csv('output.tsv', sep='\t', index=False, header=False)